Title of project: Genetic Transformation and Development of Elite Transgenic Maize (Zea mays L.) for Biotic and Abiotic Stresses Tolerance
Project funding: Externally funded (NASF)
Total approved cost of the project (for IIMR): 154.67 lakh
Objective:
- Development of novel transformation constructs for herbicide tolerance and phosphorus use efficiency.
- Transformation of maize for herbicide tolerance and insect resistance.
- Molecular analysis, phenotyping and bioassay of transgenic lines.
- Conversion of elite maize inbred with transgenes and pyramiding of transgenes in the elite inbred backgrounds.
Project team:
- PI: Dr. Pranjal Yadava (1.07.2015 – 04-01-2017), ICAR- Indian Institute
- of Maize Research, New Delhi
- Dr. Tanushri Kaul (5.01.2017– 31.12.2018), International Centre for Genetic Engineering and Biotechnology, New Delhi.
- CC-PI: Dr. Tanushri Kaul (1.07.2015 – 04-01-2017), International Centre for Genetic Engineering and Biotechnology, New Delhi.
- Dr. Krishan Kumar (5.01.2017– 31.12.2018), ICAR- Indian Institute of Maize Research, New Delhi
- CC-PI: Dr. Rakesh Bhowmick, ICAR-Vivekananda Parvatiya Krishi Anusandhan Sansthan, Almora
- CCo-PI: Dr. Ishwar Singh, ICAR- Indian Institute of Maize Research, New Delhi
Major achievements: – IIMR
Identification of contrasting genotypes under low-phosphate stress in maize:
Optimized concentrations of various nutrients in Hoagland’s solution for hydroponics-based method of screening for low-phosphate stress tolerance in maize. Screened 40 maize inbred lines for phosphate stress/PUE under hydroponic condition and identified highly tolerant (BML5) and highly susceptible (BML10) lines (Figure 1) which could be utilized in breeding programme for developing PUE hybrids and to study molecular mechanisms playing role for phosphate stress tolerance.
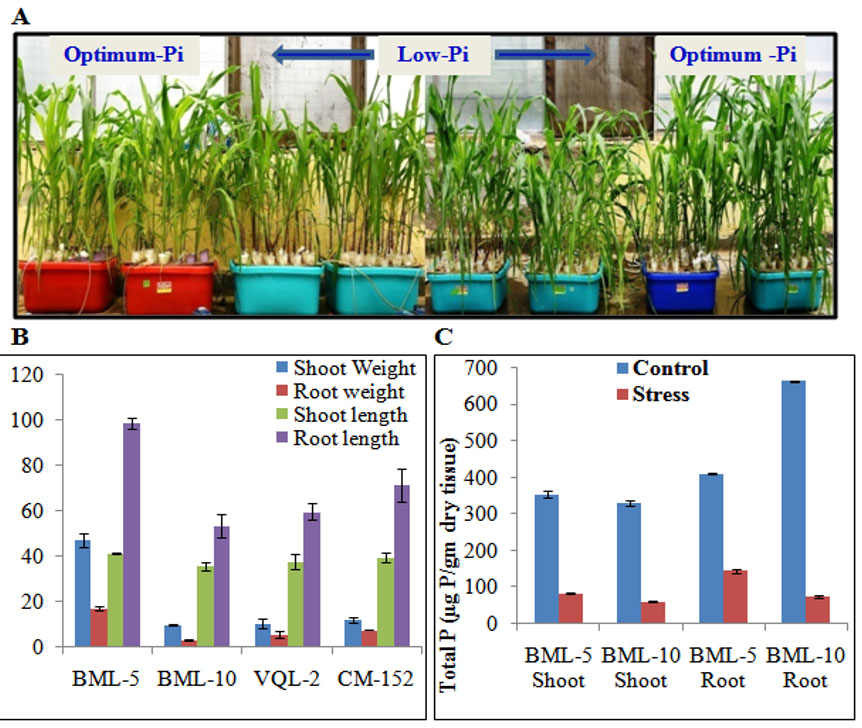

Figure 1: Screening for PUE under hydroponic culture in tropical maize.
(A) Plant growth under hydroponics in optimum and low-phosphate (Pi) conditions. (B) Shoot and root dry weight (in gm) and length (in cm) under low-phosphate stress condition. (C) Phosphate accumulation in shoot and root of high and low PUE lines. (D) BML-5 (highly tolerant) and BML-10 (highly susceptible) lines under low and optimum phosphate conditions.
Identification and expression profiling of phosphate stress responsive genes in maize: Based on in silico analysis (sequence similarity and functional domain analysis), 12 phosphate (Pi) starvation responsive key genes (five regulatory genes, two high affinity Pi transporters and four secretory proteins) in Zea mays were identified. Quantitative real-time PCR (qRT-PCR) and semi-quantitiative RT-PCR based expression analysis revealed that 11 out of 12 genes in Pi deficient condition were significantly differentially regulated in maize (Figure 2).
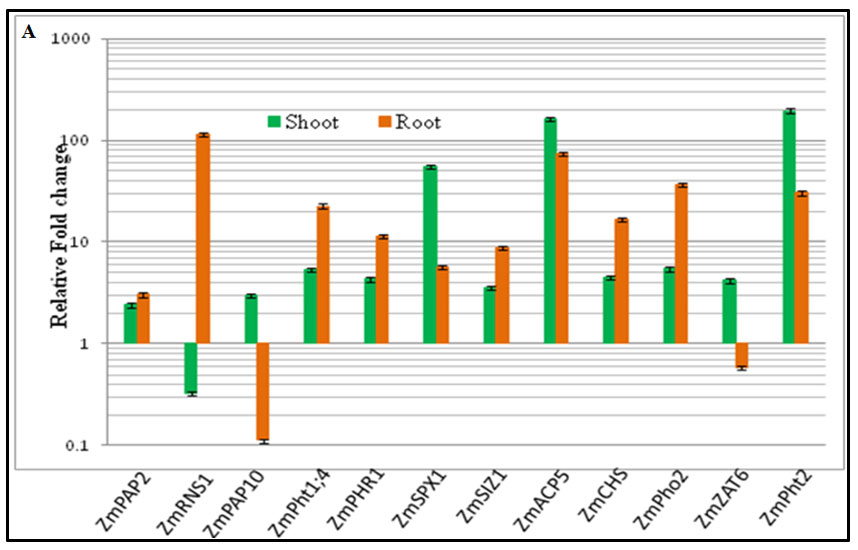
Figure 2. Expression analysis identified genes involved in phosphate responsive pathway.
The qRT-PCR based expression analyses of predicted regulatory genes [PHR1 (phosphate starvation response 1), SPX1 domain containing protein, SIZ1, Pho2 & Zinc finger domain containing TF (ZAT6)], secretory proteins [PAP10 (purple acid phosphatases), PAP7, ACP5 (acid phosphatases 5) & RNS1 (ribonuclease)] and high affinity phosphate transporters [PHT 1;1 & PHT 1;4] genes at phosphate deprived conditions in HKI-163 inbred line of maize grown under phosphate deprived (5 µM KH2PO4) (-Pi) and sufficient phosphate (+Pi) conditions (1 mM KH2PO4). Y-axis represents the relative fold change values at low phosphate condition as compared to respected genes at sufficient phosphate conditions.
Transcriptional profile of phosphate responsive miRNAs in maize:
Identified differentially expressed miRNAs under low-phosphate stress in tropical maize through RNA-seq profiling. The transcriptome analysis revealed 35 known miRNAs (12 up & 23 down-regulated) belonging to 14 families in shoot and 27 known miRNAs (15 up & 12 down-regulated) belonging to 12 families in root expressed differentially under low-phosphate stress. Among these, 10 miRNAs (viz., miR827, miR399 members, miR156 members, miR159 member etc.) were common in roots and shoot/leaf tissue. We also identified 40 (18 up & 22 down-regulated) and 46 (37 up & 9 down-regulated) novel miRNAs in shoots and roots respectively. These highly dysregulated miRNAs may be a key factor affecting tolerance of maize plant in scarce phosphate condition.

